Genome and population dynamics during chronic infection with Helicobacter pylori
Lorem ipsum dolor sit amet, consetetur sadipscing elitr
The infection with the stomach bacterium Helicobacter pylori is one of the most common infections of human. More than one half of the world population is infected with this pathogen. H. pylori infection is frequently acquired in childhood and persists for decades, such that H. pylori is a particularly characteristic example of a chronic infection, the overarching theme of this collaborative research center. Not all H. pylori infected persons suffer from consequences of the infection: approximately 85 percent will never develop any symptoms. Ten to fifteen percent of infected individuals will develop either stomach ulcers or ulcers of the duodenum. One percent will develop the most dangerous complication of H. pylori infection, stomach cancer. Due to the high number of infected persons, H. pylori causes about 800,000 new cases of stomach cancer per year.
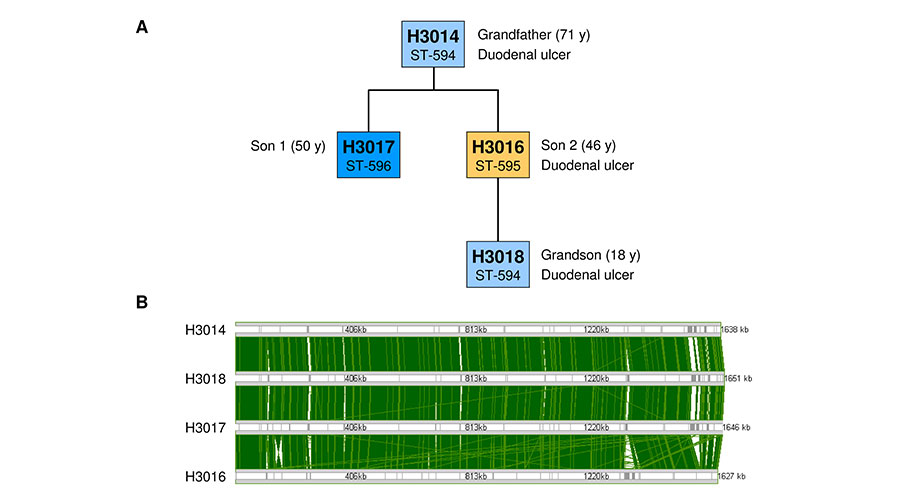
It has been known for more than 30 years, that H. pylori bacteria from unrelated patients differ from each other. Every patient carries his/her own individual H. pylori strain, or even multiple strains. H. pylori diversity is thus similar to that of human fingerprints.
The research group around Sebastian Suerbaum aims at elucidating the molecular mechanism that cause the extremely high genetic diversity of H. pylori. They want to clarify how the high variability of these bacteria contributes to the course of H. pylori infection.
Scientific work programme
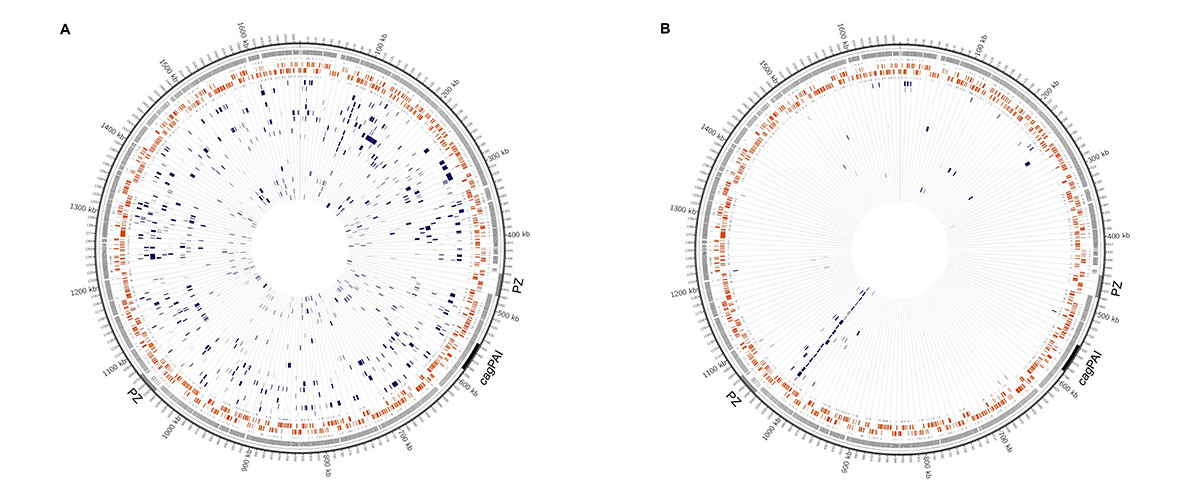
The research group uses different high throughput sequencing technologies (Next Generation Sequencing) to decipher H. pylori genome sequences. To understand the mechanism that generate genetic diversity, we sequence the isolates from diverse patients, and from different areas of the stomach of one individual.
Furthermore, we study isolates cultures from the same human individual at different time points several years apart, with the aim to study whether the bacteria adapt to their individual human carrier. The researchers use the Illumina MiSeq technology for the analysis of large numbers of H. pylori isolates. This technology usually assembles the genomes in an average of 25 fragments (contigs). The Single Molecule, Real-Time (SMRT) sequencing technology (Pacific Biosciences) is used to generate complete (finished) genome sequences, where the complete circular chromosome is sequenced as one continuous sequence. Due to the higher cost of this technology, only selected strains can be sequenced by SMRT technology, and a combination of both methods permits optimal throughput and a sufficient number of finished reference genomes. Genome sequencing is combined with experimental genetic and biochemical research approaches. We construct mutants where selected H. pylori genes have been inactivated, and study the effect of such mutations on the ability of H. pylori to generate genetic diversity.
This will enable the elucidation of the molecular mechanisms generating mutations in H. pylori and their exchange between strains by recombination. One further topic studied in this project is the epigenetics of H. pylori. SMRT sequencing permits to identify bases in the bacterial genome that have undergone modification by methylation. H. pylori change their methylation pattern during chronic infection, and the research group studies the consequences of such methylation changes for the infection.
Lorem ipsum dolor sit amet, consetetur sadipscing elitr
Sebastian Suerbaum is talking about his research at CRC 900
Sebastian Suerbaum has been part of the CRC since it was founded in 2010. His project A1 focuses on investigations into the genetic variability of H. pylori and the link between this genetic diversity and the course of chronic infections. Together with Professor Tümmler Professor Suerbaum also leads project Z1, which offers high-throughput sequencing and bioinformatics as a service for the whole CRC. Professor Suerbaum is also vice-speaker of the CRC.
Publications of the project A1
Contact

Prof. Sebastian Suerbaum
Chairman, Dept. of Medical Microbiology and Hospital Epidemiology
Max von Pettenkofer Institute
Ludwig-Maximilians-Universität München
Pettenkoferstr. 9a
80336 München
+49 89 2180-72800
suerbaum@mvp.uni-muenchen.de