Chronic Pseudomonas aeruginosa infections in cystic fibrosis: pathogen microevolution and host genetic predisposition
Pseudomonas aeruginosa grown on blood agar
Patients suffering from cystic fibrosis (CF) are affected by dysfunctions of exocrine glands. The defect or limited function of a certain transporter protein results in aberrantly composed secretions produced in the guts, stomach and pancreas and also in the airways. The secretions are more viscous and impair the function of the affected organs. In the airways of CF patients, mucosal tissue is not sufficiently supplied with water and thus more prone to bacterial infection. Airways of 80 % of adult patients are permanently colonised by the bacterium P. aeruginosa. The bacteria cannot be cleared by the immune system of CF patients and establish a chronic infection accompanied by increasing damage of the airway tissue and drastic reduction of the lung function. The chronic P. aeruginosa infection thereby determines the clinical status and the further course of the disease for 90 % of the CF patients overall.
The aim of this project is to analyse the contribution of genome variations of the pathogens as well as of the human hosts to the severity of the disease courses, which can vary significantly from between patients. For P. aeruginosa, characteristics of the initially colonising strains are involved, but also genome variations introduced during the chronic infection. This microevolution within the CF airways can modify bacterial phenotypes and apparently contributes to the adaption of the bacterial lifestyle to the CF lung habitat, to increased resistance against mechanisms of the host immune system and also against antimicrobial treatment. Therefore, the long-term objective of the project is to characterize genomic variation between the hosts and the bacterial microevolution during the courses of the chronic infection, and to look for eventual associations between them. The identification of CF-specific bacterial microevolution events and consequential habitat adaptation strategies could reveal targets for new treatment strategies for chronically infected CF patients.
Scientific work programme
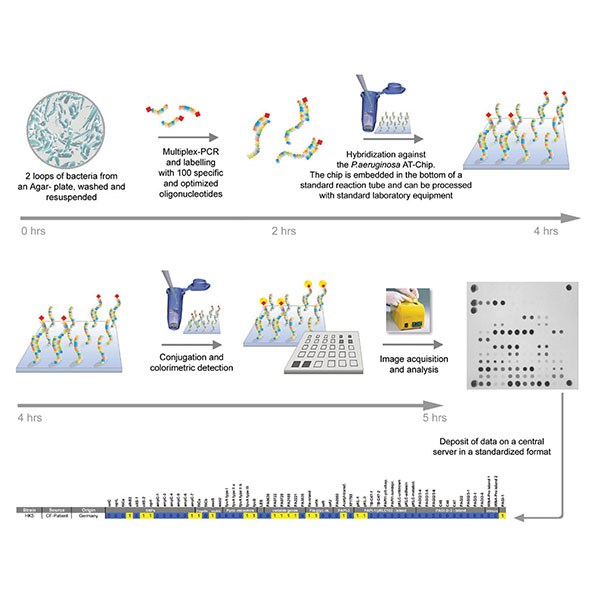
For the detailed evaluation of the microevolution and adaptation processes of P. aeruginosa to the CF lung habitat a unique strain collection held at Hannover Medical School is utilised. The collection comprises sequential P. aeruginosa isolates obtained from the airways of CF patients in half year intervals from the onset of the chronic colonisation on. These sets of sequential isolates allow a long-term view on the microevolution process over the whole courses of disease. Disease courses with different grades of severity are represented, and for some cases the sets of isolates cover time spans of more than 25 years.
For this project twelve sets of sequential isolates were selected, from the six most severely affected patients and the six patients with the mildest clinical courses. For each of these twelve courses, isolates taken in one year intervals were chosen for detailed analyses, as long as the initially colonising P. aeruginosa clonal lineage could be detected in the airways. These subsets of isolates were analysed by NGS genome sequencing and phenotypic characterisation.
The NGS data was screened for genome variations (nucleotide exchanges, small indels, larger deletions, uptake of new DNA) that occurred after the initial colonisation of the CF lungs, and by comparison over all datasets microevolution “hotspots”, genomic loci with increased mutation frequencies, could be identified which are apparently associated with diversification and adaptation processes in the CF lung habitat.
In addition, phylogenetic trees (based on shared or individual occurrences of the detected genome variations) were constructed for each of the twelve sets of sequential isolates which display the respective modes of microevolution. Overall, the detected genome variations, their presumed effects and the detected microevolution modes are questioned for associations with the severity of the patients’ clinical courses and for the occurrence of common CF-specific traits.
The twelve subsets of sequential isolates are also analysed by competitive fitness experiments. Studying the competitive growth of the isolates in nutrient-rich and nutrient-poor medium shall reveal the abilities of the bacteria to thrive under these conditions after different time spans of adaptation to the CF lung habitat. The results should also display the influence of adaptation and specialisation to an unusual host under continuous pressure of the immune system on the global fitness of P. aeruginosa and on their ability to thrive afterwards in other environments.
For the fitness experiments the respective sequential isolates are cultivated together in liquid culture, and samples are taken at different time points. The portions of the individual isolates are determined from amplicon sequencing data by utilisation of isolate-specific trains detected by the previous genome sequencing as markers.
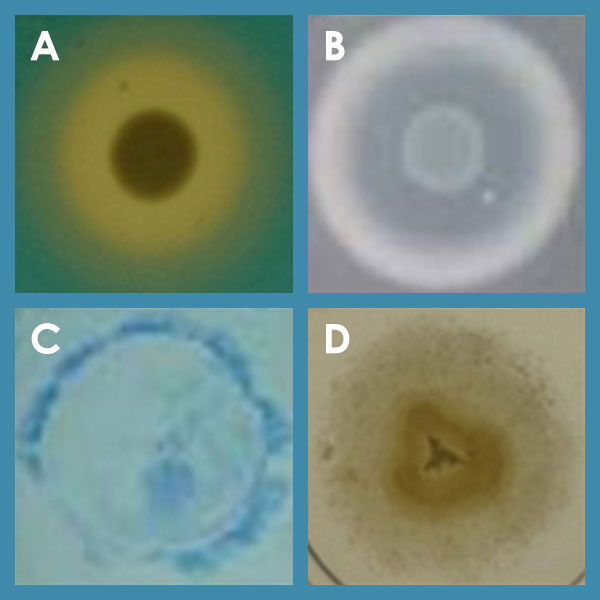
Various phenotypes of P. aeruginosa; A: siderophore secretion; B: protease secretion; C: twitching motility; D: swimming motility
Two further questions were also addressed by bacterial NGS sequencing within this project, i. e. the intraclonal diversity of P. aeruginosa and, based on the sequencing results for the sequential isolates, the occurrence of the microevolution events arisen during the chronic CF lung colonisation in P. aeruginosa isolates from other sources. For studying the intraclonal diversity 100 isolates from different sources representing the globally most prominent clonal lineages C40A and PA14 were genome sequenced, screened for common and individual sequence variations, and selected isolates were also compared for competitive fitness.
The second topic was addressed by choosing certain genomic loci for an amplicon sequencing approach. Mutation hotspots known from the results for the sequential isolates and genes described in the literature as being involved in CF lung adaptation were sequenced in other P. aeruginosa collections to display the global mutation spectrum in these >30 loci and to describe mutations as either global or habitat-specific.
These collections contained P. aeruginosa CF isolates from other hospitals as well as isolates from other habitats (isolates from COPD patients, from acute infections, environmental isolates). Strains displaying notable mutations shall be examined individually afterwards for fitness traits and for changes in protein functions encoded by the affected genes and subsequent phenotypic changes.
The influence of the genetic background of the CF patient on the course of the chronic P. aeruginosa infection shall be analysed by examining the genomes of the twelve patients from which the sequential bacterial isolates have been chosen for detailed analysis. Genome variations associated with either the mild or the severe clinical courses are candidates for potential modifiers of the severity of the chronic infection and shall be tested for occurrence in other CF patients. The modulator candidates shall be screened in large patient cohorts for whom DNA has been deposited in the European-Canadian unified CF DNA biobank storing samples of several thousand patients from Germany, Switzerland and Canada. Modifiers confirmed in the large patient cohorts would then represent novel biomarkers for the severity of chronic P. aeruginosa infection in CF.
Burkhard Tümmler ist talking about his research at CRC 900
Burkhard Tümmler has been part of the CRC since it was founded in 2010. His project A2 investigates the evolution of Pseudomonas aeruginosa during chronic infections in patients with cystic fibrosis. Together with Professor Suerbaum Professor Tümmler also leads project Z1, which offers high-throughput sequencing and bioinformatics as a service for the whole CRC.
Publications of the project A2
Contact

Prof. Burkhard Tümmler
Clinical Research Group – OE6711
Clinic for Pediatric Pneumology, Allergology und Neonatology
Hannover Medical School
Carl-Neuberg-Str. 1
30625 Hannover
+49 511 532-2920
+49 511 532-6723
Tuemmler.Burkhard@mh-hannover.de