Novel host modulatory factors of Helicobacter species
Chronic inflammatory bacterial infections are rare, are difficult to detect, and can be associated with severe diseases including cancer. Therapeutic and prophylactic measures are not easy to implement against these infectious diseases, since their traits include various activities to evading and manipulating the immune system of the host organism. One paradigm and model organism for chronic inflammatory disease is the bacterial agent of severe human stomach pathologies, Helicobacter pylori. H. pylori is predominantly acquired in early childhood within families and, even at an early age, can be linked to serious symptoms such as anemia, growth retardation, and perforating gastric ulcers. With increasing age, the incidence of gastric and duodenal ulcers rises in infected persons, and the infection can also promote various malignancies of the stomach. With a global view on public health, the infection is one of the most prevalent, since about 50% of the world population carries the bacteria. Every single infected individual has their own individually adapted H. pylori strain, since the bacteria can quickly diversify and adapt upon transmission. A spontaneous clearance of the infection has been observed quite rarely. Simple therapeutic options are not available yet, nor is a prophylactic vaccine. Closely related bacteria exist in humans and various animals, where they can cause chronic infections and inflammation in the gastrointestinal tract. The biology of these infectious agents is therefore of broad interest for understanding chronic bacterial infections and devising new ways to antagonize them.
Scientific work programme
The research group of Christine Josenhans is studying the mechanisms that enable Helicobacter pylori and related organisms to undermine the immune system of the host and thereby to promote the establishment of a chronic-active infection. In this project place, researchers in her lab place particular focus on a virulence module of the bacteria, the genomic pathogenicity island CagPAI. The genetic information of this island generates a complex transport system in the bacterial envelope, which enables the bacteria to gain access to the host cell and influence cell signaling. These signals activate for instance a chronic inflammatory response and promote the persistent colonization of the human stomach mucosa. The presence of the CagPAI is also strongly associated with a higher risk of gastric cancer in H. pylori-infected individuals.
The Josenhans lab investigates selected protein-protein interactions of CagPAI proteins with each other and with host cell receptors. In addition, they strive to clarify which factors are transported through the system to affect host cells and which host factors or receptors are addressed in turn. The studies also provide information about target points in the bacteria which may instruct novel therapies and improve vaccination strategies.
For the clarification of these mechanisms, they apply numerous biochemical, immunological and cell biology methods. In addition, sophisticated infection models have been established. One specific approach is based on evolutionary genetic and postgenomic analyses. These analyses include using partial or whole genome sequences of these very variable bacteria and combine them with mathematical models which summarize the global variation and selective forces on genes which encode proteins involved in host interaction. In this manner, detailed information on predicted in-host selective forces and protein interactions on the bacterial surface and between host and bacteria can be gathered. This data is being used to gain deeper insight into the stability and environmental molding of host-pathogen interaction interfaces.
Furthermore, biochemical and microscopic methods are used which visualize the metabolic or inflammatory adaptation of the bacteria to the variable and fluctuating situation in the host environment.
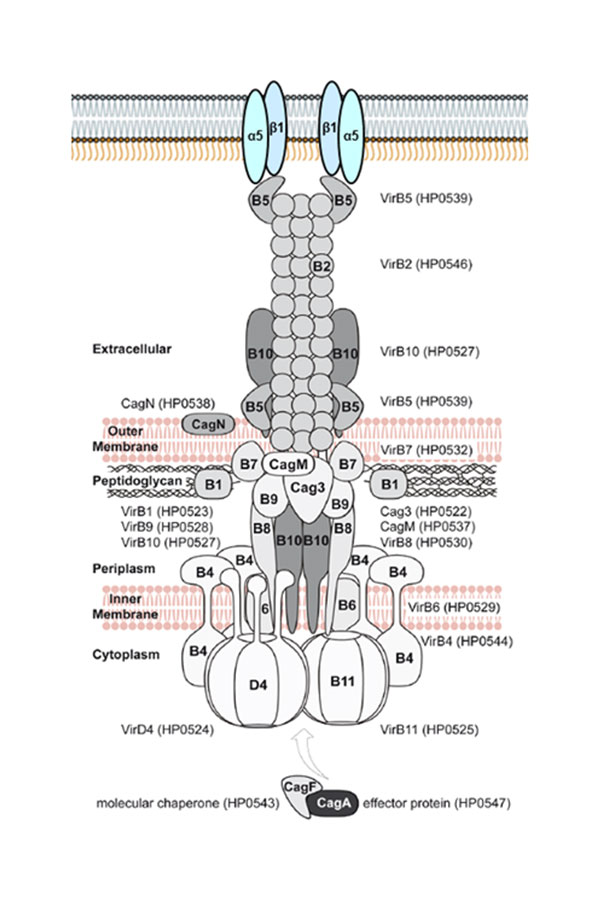
Model of the Cag t4ss of H. pylori, highlighting diversifying selection on outer and secreted components of the t4ss apparatus. (Olbermann et al., PLoS Genetics, 2010)
Christine Josenhans is presenting her research at CRC 900
Christine Josenhans has been part of CRC since it was founded in 2010 and with her team researches the mechanisms allowing Helicobacter pylori and related bacteria to cause long-term infections.
Publications of the project B6
Contact

Prof. Dr. rer. nat. Christine Josenhans
Dept. of Medical Microbiology and Hospital Epidemiology
Max von Pettenkofer Institute
Ludwig Maximilians Universität München
Pettenkoferstrasse 9a
80336 München
+49 8921 8072 826
josenhans@mvp.uni-muenchen.de
